Note: The
taxmodule was developed with Titus Brown, Taylor Reiter, Luiz Irber, and Hannah Houts. The approaches described have emerged over years ofsourmashdevelopment and exploration by these authors as well as Phil Brooks, Bry Moore, Mo Abuelanin, and a number of other keysourmashcontributors. We’re preparing preprints to properly describe this functionality, but for now please engage with us on gitter, github, twitter, etc!
With sourmash v4.2, we introduced a new module, sourmash tax (alias: taxonomy) for integrating taxonomic information into sourmash gather results. With this addition, the sourmash tax approach supercedes the sourmash lca approach as our recommended method for taxonomic integration.
sourmash gather finds the minimal set of reference genomes that match your query
The approach described here relies on sourmash’s Scaled Minhash sketches, which are comprised of a chosen proportion of k-mers from an input sequence dataset, rather than a static chosen number of k-mers, as in standard MinHash. While this means Scaled MinHash sketch size scales with dataset size, downsampling sequencing datasets in this way enables estimation of containment, which facilitates comparisons between datasets of very different sizes (e.g. trying to figure out which genomes are in your metagenome). You can find a bit more on Scaled MinHash in our sourmash use cases paper (2019).
sourmash gather is an approach that uses containment to find the smallest set of reference genomes that contain all k-mers in your query sketch. Titus gave an excellent talk on sourmash gather recently (slides), and the approach is described in detail in Luiz Irber’s dissertation. Luiz’s benchmarking suggests that gather is a very accurate method for doing strain-level resolution of genomes, but I’ll leave a full description for our preprint (hopefully out by year-end :).
As the exact strains from novel sequencing projects are often not present in reference databases, gather results commonly include matches to multiple strains of the same species, if there are multiple strains present in the reference database. These matches represent the portions of our novel query strain(s) that share genomic content with strains in the reference database.
sourmash tax annotates gather results and conducts LCA summarization
To connect to all the research that has been done to describe organisms at the species level (and above), we wanted to be able to integrate taxonomic lineage information to the gather-reported genome matches. Enter sourmash tax, which adds taxonomic integration and optionally uses a modified lowest common ancestor (LCA) approach to summarize results to higher taxonomic ranks.
How is this different from current k-mer LCA methods?
The main innovation of the gather–>tax method is that LCA aggregation occurs at the genome level, or really at the “groups of k-mers” level, rather than at the individual k-mer level.
Single k-mers, either for true biological reasons or as a result of reference inaccuracies, can sometimes match to quite different organisms and derail the individual k-mer LCA approach (nicely laid out in this twitter thread):
Consider the case where you have 20 hits, 19 of which are to the Genus Prevotella, and 1 of which is to the Archaea Methanobrevibacter.
— Mick W@tson (@BioMickWatson) July 30, 2021
Most people would say "Prevotella!" but the lowest common ancestor would be "cellular organisms" - this is not useful
In our experience, aggregating k-mer matches to the set of best genome matches with gather prior to LCA means contamination and other reference issues are far less likely to impact LCA annotations. As an added benefit, this approach allows us to do taxonomic assignment with multiple taxonomies (e.g. GTDB if available, NCBI for the remaining), so long as the gather was run against the appropriate databases.
How does it work?
This approach relies upon the fact that gather reports both the total fraction of the query matched to each reference genome, as well as a non-overlapping f_unique_to_query, which is the fraction of the query uniquely matched to each reference genome. The f_unique_to_query for any reference match will always be between 0 (0% of query matched) and 1 (100% of query matched). For query k-mers that match more than one genome, gather iteratively assigns these k-mers to their maximal (best containment) genome match in order to report the non-overlapping f_unique_to_query.
For a query matched to multiple references, the f_unique_to_query of all of its reference matches will sum to at most 1 (100% of query matched). We use this property to do LCA aggregation of gather results. For example, if the gather results for a metagenome include matches to 30 different strains of a given species, we can sum the fraction uniquely matched to each strain to obtain the total fraction uniquely matched to this species.
Note that gather results themselves are still affected by the completeness of the reference database, as are all reference-based analyses. Long DNA k-mers are quite specific, so if you get a match, it’s pretty reliable. However, if you don’t get a match, it may mean that the species or genus you’re querying may not be closely related enough to anything in the reference database. We’ve been exploring protein k-mers to overcome some of these challenges and enable matching across larger evolutionary distances. I’ll go into detail in my next blog post!
The sourmash tax commands
Two commands in sourmash tax use our LCA approach: metagenome and genome. We hope the intended input query for each of these methods is self-explanatory :).
For all methods, you’ll need to have sketched your query or queries, and run sourmash gather against a reference database. You can find pre-prepared GTDB reference databases here. Second, you will need a file containing the taxonomic information for all genomes in your reference database (gtdb-rs202 file here).
sourmash tax metagenome summarizes gather results for each query metagenome by taxonomic lineage, and (by default) reports the results at each taxonomic rank.
sourmash tax genome reads gather results for each query genome and attempts to classify to the taxonomic lineage of the best gather result. Our current approach uses a threshold of 10% query coverage before assigning a classification, meaning that if an insufficient portion of our query is matched at the species level, we will use the LCA lineage summarization approach above to assess the match at each rank, and classify to the rank where our classification threshold is met. Alternatively, classification can be conducted at a specific taxonomic rank, regardless of classification threshold. This method is our newest addition, and we welcome feedback and results to help improve it going forward.
Note we also offer krona output from metagenome or rank-classified genome results, as well as a lineage_summary output for tax metagenome that may be useful for external multi-sample visualization tools.
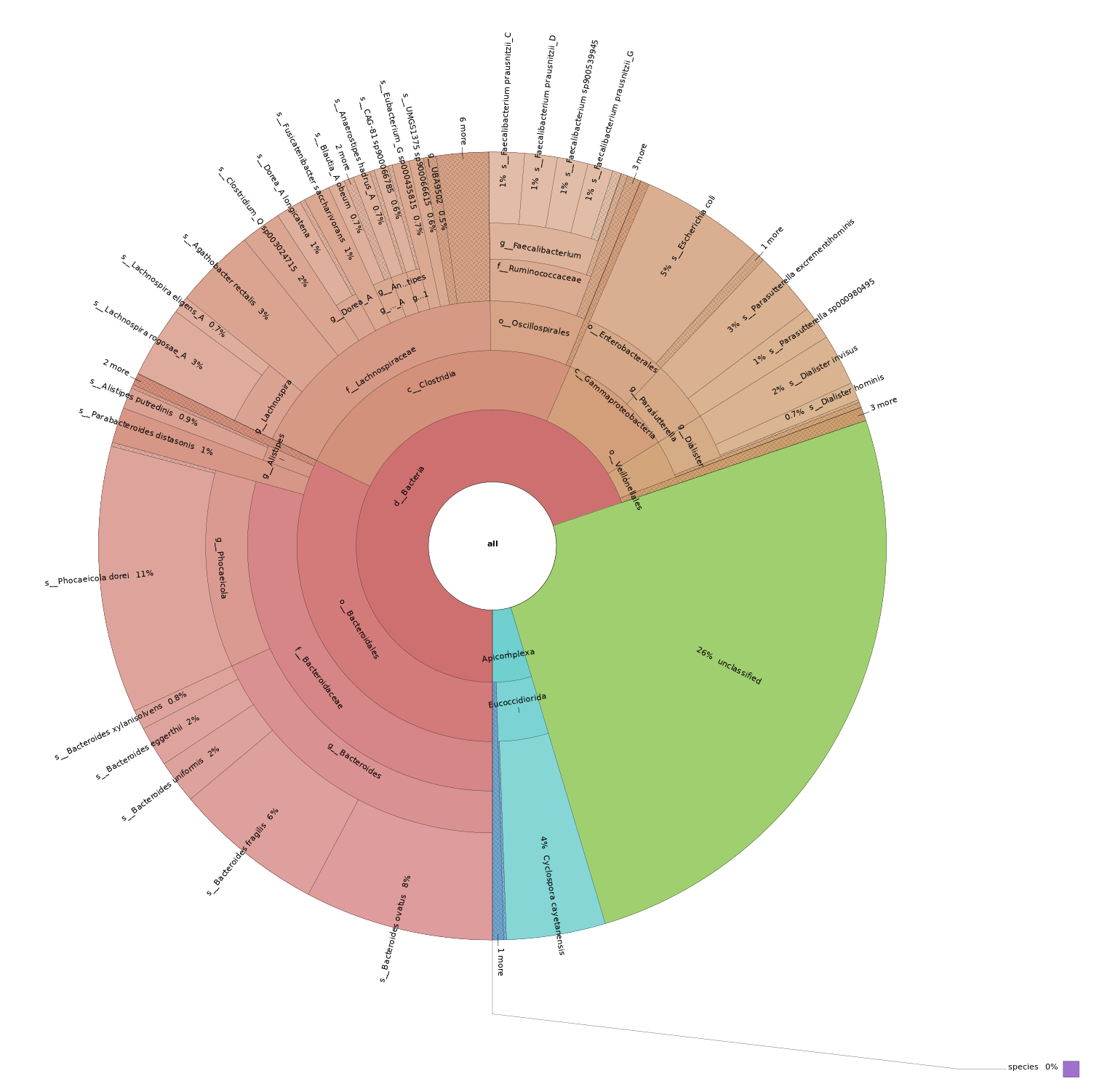
There are two additional commands within the sourmash tax submodule, prepare, and annotate.
tax prepare is a method for converting a csv of taxonomic lineage information into an sqlite database to enable faster loading and lineage assignment. It can also be used to combine lineage information for more that one database (e.g. GTDB, NCBI).
tax annotate annotates gather results with taxonomic information, without doing any LCA summarization.
What about the sourmash lca methods?
The sourmash lca submodule uses the LCA approach pioneered by Kraken and used by many other tools. It uses taxonomic information to classify individual k-mers, and then infers taxonomic distributions of metagenome contents from the presence of these individual k-mers. The lca subcommands work with LCA databases, which contain taxonomic information by design, and are pretty neat for other reasons too.
While we recommend using the gather–> tax method, the lca module isn’t going anywhere, and many may still find it interesting or useful.
Additional Info
Please see the documentation for additional details, and contact us on gitter with questions and comments, or file an issue on our github. We’d love to hear if and how these methods are useful to you!